|
flavour
|
|
flavour
|
The program produces figures presenting 68%, 95% and 99% CL allowed regions in parameter space. To wit, we represent regions where the specific BGL model is able to fit the imposed experimental information at least as well as the corresponding goodness levels. Some comments are in order. This procedure corresponds to the profile likelihood method. In brief, for a model with parameters 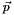 , we compute the predictions for the considered set of observables
, we compute the predictions for the considered set of observables 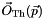 . Then, using the experimental information
. Then, using the experimental information 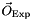 available for those observables, we build a likelihood function
available for those observables, we build a likelihood function 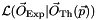 which gives the probability of obtaining the experimental results
which gives the probability of obtaining the experimental results 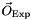 assuming that the model is correct. The likelihood function
assuming that the model is correct. The likelihood function 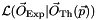 encodes all the information on how the model is able to reproduce the observed data all over parameter space. Nevertheless, the knowledge of
encodes all the information on how the model is able to reproduce the observed data all over parameter space. Nevertheless, the knowledge of 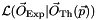 in a multidimensional parameter space can be hardly represented and one is led to the problem of reducing that information to one or two-dimensional subspaces. In the profile likelihood method, for each point in the chosen subspace, the highest likelihood over the complementary, marginalized space, is retained. Let us clarify that likelihood – or chi-squared
in a multidimensional parameter space can be hardly represented and one is led to the problem of reducing that information to one or two-dimensional subspaces. In the profile likelihood method, for each point in the chosen subspace, the highest likelihood over the complementary, marginalized space, is retained. Let us clarify that likelihood – or chi-squared 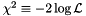 – profiles and derived regions such as the ones we represent, are thus insensitive to the size of the space over which one marginalizes; this would not be the case in a Bayesian analysis, where an integration over the marginalized space is involved. The profile likelihood method seems adequate to our purpose, which is none other than exploring where in parameter space are the different BGL models able to satisfy experimental constraints, without weighting in eventual fine tunings of the models or parameter space volumes. For the numerical computations the libraries GiNaC and ROOT are used. *
– profiles and derived regions such as the ones we represent, are thus insensitive to the size of the space over which one marginalizes; this would not be the case in a Bayesian analysis, where an integration over the marginalized space is involved. The profile likelihood method seems adequate to our purpose, which is none other than exploring where in parameter space are the different BGL models able to satisfy experimental constraints, without weighting in eventual fine tunings of the models or parameter space volumes. For the numerical computations the libraries GiNaC and ROOT are used. *